“What I really like in this lab is the creative ways in which problems are solved and methods improved. It’s fun to solve such puzzles together to develop new solutions that in turn lead to exciting new discoveries.” ~ Naomi Botay
“Our lab has a collaborative environment, among each other or with other labs. This results in fruitful discussions about which screening to do next or how to upgrade existing methods even more.” ~ Esther Uijttewaal
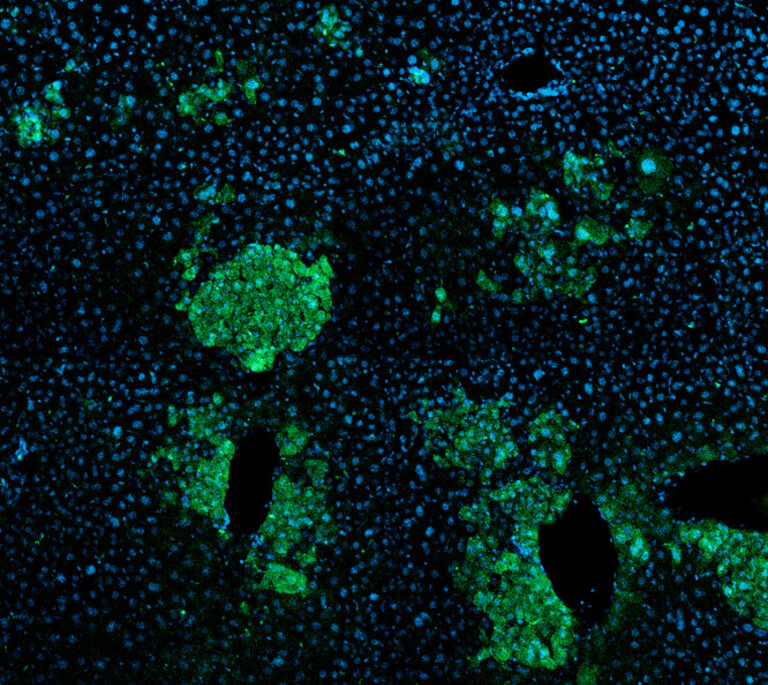
“I’m excited to be working on CRISPR screens to investigate the genetic dependencies of cancer cells. With an unbiased and high-throughput approach like this, we might be able to uncover new vulnerabilities that could be used to specifically target cancer cells.” ~ Sophie van der Leij
“This project triggers my interest in performing cutting-edge research and the further development and improvement of CRISPR methods. Using complex and innovative technologies to be able to improve the understanding of a wide variety of biological processes in a healthy or diseased state motivates me to come to the lab every day.” ~ Esther Ujittewaal
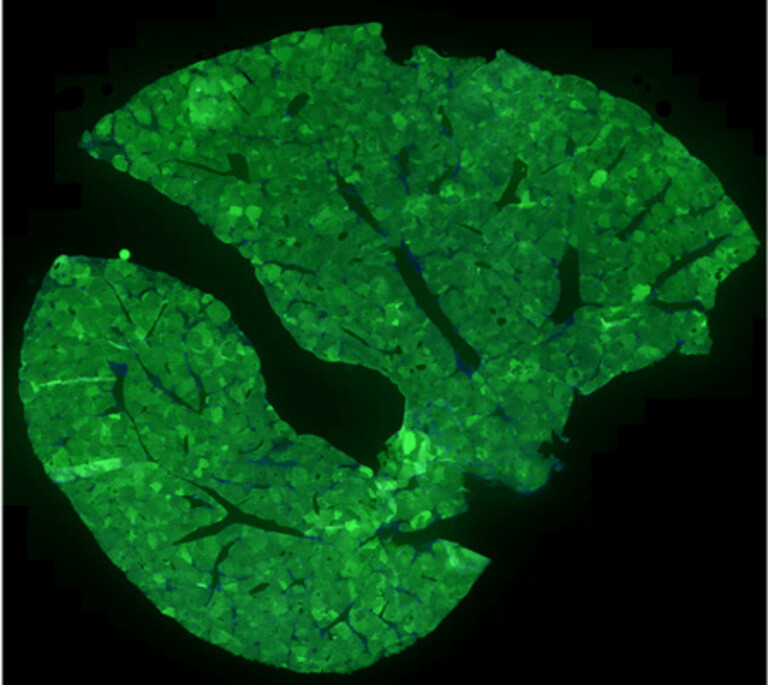
“I always wanted to study biology in a real context, and this project is exactly my dream project. On top of the unbiased genetic screening in a living organ, developing new methods and testing possibilities are real fun and make me work enthusiastically. Our group work as a team to create synergy, and this is what I am doing by adding colours to CRISPR-StAR and make it even more powerful..” ~ Joonsun Lee
“I am very fascinated by the possibilities coming along with CRISPR-Cas9. Applying and improving this system is very exciting to me and I am happy to have landed in a lab with such great expertise on this specific topic. Digging into the various methods for preparation and analysis of our screens also has been and will remain an interesting challenge throughout my project.” ~ Tobias Potzler
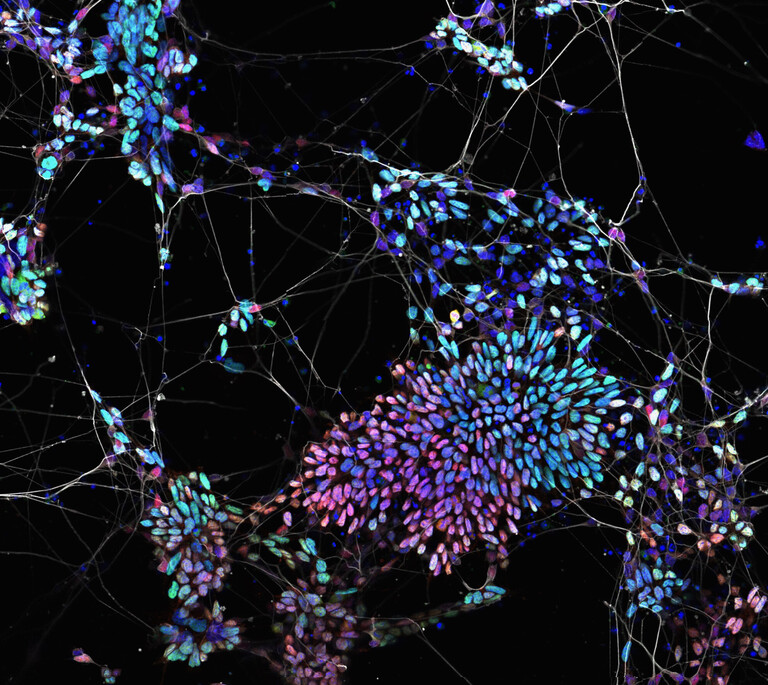
“I am always surprised how many unique approaches to the current difficult scientific problems we are tackling; let it be exciting questions in lineage transitions or bringing new methods circumventing limitations in CRISPR-Cas9 screening. For me, working in this lab quickly developed ‘outside of the box’ thinking, gave a steep learning curve in adapting numerous methods and allowed asking deeper questions from the data.” ~ Gintautas Vainorius