Histone variants shape the Arabidopsis epigenome
DNA can be transcriptionally active or silent, depending on its state of packaging into chromatin. The “packaging proteins”, called histones, exist in many variants. A new study now shows that these histone variants are key drivers to determine whether chromatin is in a transcriptionally active state – or not. The study, by the group of Fred Berger at the Gregor Mendel Institute of Molecular Plant Biology, was published in the journal eLife on July 19.
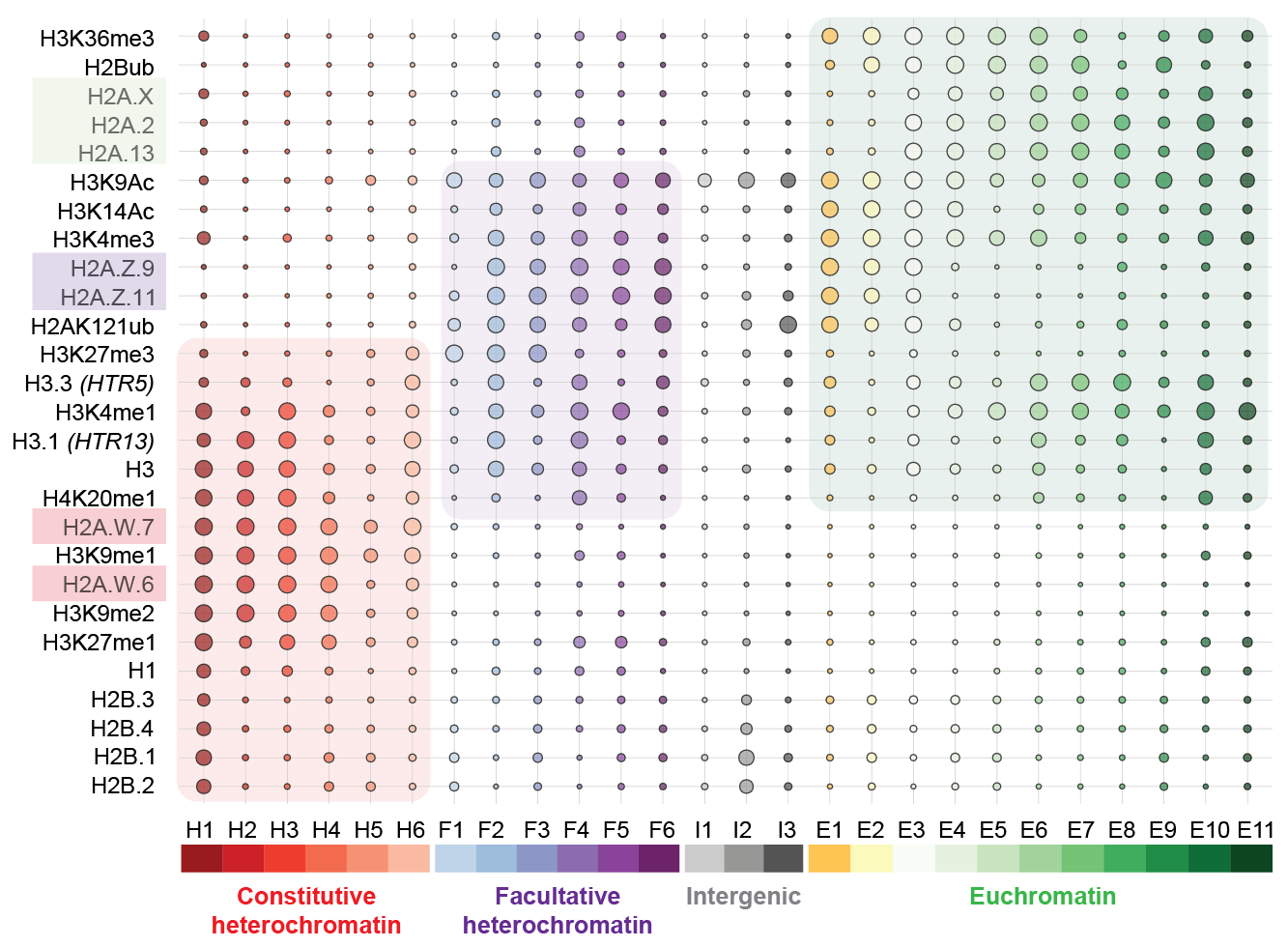
DNA does not float freely within the nucleus; instead, eukaryotic genomes are packaged into chromatin, a structure in which DNA is wrapped tightly around proteins called histones. Whether or not DNA can be “read” and translated into proteins depends also on its packaging into chromatin. Three types of packaging exist: euchromatin, in which genes are active, facultative heterochromatin, in which genes are repressed, and constitutive heterochromatin, containing transposons.
Histones can be modified by so-called posttranslational modifications, such as methylation, acetylation, etc. Scientists have so far studied the relationship between posttranslational modifications and chromatin states. However, histones also exist in many variants – particularly in flowering plants, histones have diversified into a range of differing variants. How these histone variants contribute to chromatin states in eukaryotes was not known yet.
In a new study, the group of Frederic Berger, senior scientist at the Gregor Mendel Institute of Molecular Plant Biology (GMI) of the Austrian Academy of Sciences, studied histone variants and posttranslational modifications to find which factors drive whether chromatin is in euchromatic or heterochromatic state. Published in eLife, the study seeks to delineate the relationship between histone variants, posttranslational modifications, and chromatin states in the model flowering plant Arabidopsis thaliana.
“The strength of this study lies in using orthogonal approaches – bioinformatics, biochemistry, and genetics – to tackle the question of how histone variants and posttranslational modifications drive chromatin state”, Berger explains.
The team, including former PhD student Bhagyshree Jamge, staff scientist Zdravko Lorkovic and bioinformatician Elin Axelsson-Ekker as first authors, found that histone variants are as important for driving the chromatin states as posttranslational modifications are. Among four core histones, H2A variants appeared to be key factors that define euchromatin, facultative heterochromatin, and constitutive heterochromatin. “In a hierarchy of importance, the H2A variants, and only then the posttranslational modifications, set up in which of these three states chromatin is. The relative enrichment of other histone variants and modifications further defines subdomains of chromatin states”, Berger adds.
These predictions from bioinformatics analyses were backed up by observing what happens when the ordered system of histone variants is perturbed. The researchers studied mutant plants in which the deposition of H2A variants was deregulated and found that chromatin states severely changed. “In these mutants, the chromatin states are scrambled: Non-sense chromatin states, that don’t exist in nature, form and naturally existing chromatin states are found in regions where they shouldn’t be, so that, for example, transposons become expressed when they shouldn’t normally be”, Berger says.
How crucial histone variants are for forming the correct chromatin state was previously not known. “This is the first time that histone variants have been assessed systematically in eukaryotes. We have no reason to assume that what we found is not valid in other eukaryotes – including in mammals, which have a similar diversification in histone variants as flowering plants”, Berger concludes. In the future, Berger and his group will assess how exactly histone variants and posttranslational modifications combinations affect gene expression. The current findings contribute mainly to plant scientist’s toolbox: “With our published data, other scientists can assess in which chromatin state a gene of interest is.”
Original publication
Bhagyshree Jamge, Zdravko J Lorković, Elin Axelsson, Akihisa Osakabe, Vikas Shukla, Ramesh Yelagandula, Svetlana Akimcheva, Annika Luisa Kuehn, Frédéric Berger (2023) Histone variants shape chromatin states in Arabidopsis eLife 12:RP87714. DOI: 10.7554/eLife.87714