A CRISPR-based lab tool for lineage tracing in vivo
During the development of multicellular organisms, individual cells give rise to multiple cell lineages, complex tissue structures, and organs. Tracing cell lineages from the zygote through early embryonic development and into more advanced developmental stages still poses a challenge to this day. Researchers have developed various techniques to lineage-trace cells during development, yet all these techniques have limitations. Plants provide an advantage that makes lineage tracing easier: plant cells are usually immobile and new cells share a cell wall with their mother cells, thus keeping them physically connected.
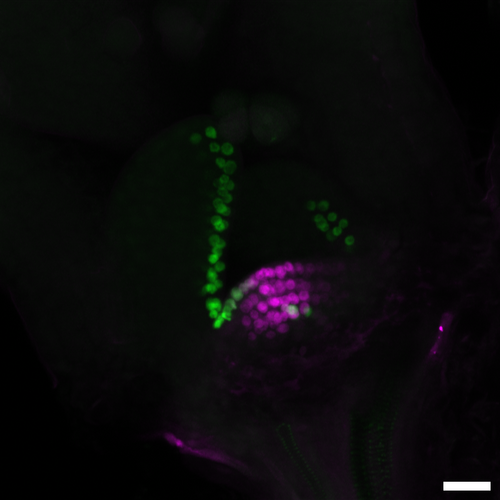
Now, GMI researchers led by postdoc Mattia Donà in the lab of Ortrun Mittelsten Scheid and in collaboration with the lab of Liam Dolan present a CRISPR-based method for cell lineage tracing in different tissues and organs, and at different developmental stages in living plants. In the study, the scientists make use of Cas9’s mutagenic power. They stably introduced a DNA sequence in the plant’s genome, in which a promoter and the coding sequence for a fluorescent protein are separated by a linker sequence containing a CRISPR/Cas9 target site. The linker is designed in such a way that it causes a frameshift in the sequence following it. Thus, the fluorophore is not produced. However, the transgene contains also the information for the Cas9 protein. Once this is expressed, Cas9 induces a double-strand break in the linker sequence, and endogenous enzymes repair this break by non-homologous end-joining, in many cases thereby restoring the frameshift. Thus, CRISPR/Cas9’s mutagenic activity “corrects” the mutation in the linker, and the fluorophore is expressed. As it is fused to a histone molecule, it is incorporated into the nucleus of the respective cells, making it glow. Once the correction is made, it cannot revert, and the strong fluorescent signal is propagated through all subsequent cell divisions. By choosing the cell type or the time of the Cas9 expression, the researchers could even choose the precise starting point of lineage tracing during development. Thus, the system allows optically tracking of cell lineages in developing plant tissues and organs in an easy-to-use, reliable, and precise fashion.
The scientists demonstrated their approach for long-term lineage tracing in two model plants: Arabidopsis thaliana and Marchantia polymorpha. Donà and his co-authors underline the easy transferability of the technique to other plants, and possibly even to other multicellular organisms.