Research
Autophagy mediated cellular quality control mechanisms in plants
Like us, plants have two types of disease states. The first one is inflicted by pathogens and fought off by the immune system. The second state, which is much less appreciated, manifests as a result of defects in the cellular proteome and organelle contents and is responded to by quality control pathways. Similar to the immune system, quality control pathways closely monitor the integrity of cellular contents and can initiate signaling cascades to either repair damaged components or remove them. In humans, shortcomings in these quality control pathways are associated with neurodegenerative diseases, such as Alzheimer’s and Parkinson’s, and aging. In plants, however, in contrast to immunity against pathogens, quality control mechanisms and their potential for improving plant productivity are mostly unexplored. Our long-term goal is to study quality control pathways at the single cell and organismal level and leverage this information to improve plant health. Currently, we are focusing on the role of selective autophagy in recycling harmful cytosolic aggregates and damaged organelles.
Selective autophagy
Macroautophagy (hereafter autophagy) is a conserved cellular quality control pathway that removes unwanted self- and non-self macromolecules to maintain homeostasis in the face of physiological and environmental fluctuations. There is a growing appreciation that autophagy is a highly selective process, with multiple layers of specificity defining the dynamics of uptake, sub-cellular trafficking, and turnover of autophagic substrates (Figure 1). However, despite these advances, the molecular details of how various autophagy cargoes and components are recognized, recruited, and recycled remain to be fully elucidated. In particular, our understanding of the molecular codes that define selective autophagy in plants is limited.
Autophagic cargo sorting involves labeling cargo with non-self tags, such as polyubiquitin chains, followed by selective engulfment into the autophagosomes. There are three key players in selective autophagy: ATG8, selective autophagy receptors, and autophagic cargoes. ATG8 directly interacts with selective autophagy receptors and labels the autophagosomes for cargo recruitment and trafficking to the vacuole. Selective autophagy receptors or cargo receptors are modular proteins that contain cargo-binding domains and conserved ATG8 interacting motifs (AIMs). This modular architecture allows cargo receptors to bring autophagic cargo to the growing autophagosome and ensures selective cargo recruitment. The autophagic receptors and cargo are then carried to the vacuole for degradation (Figure 1).
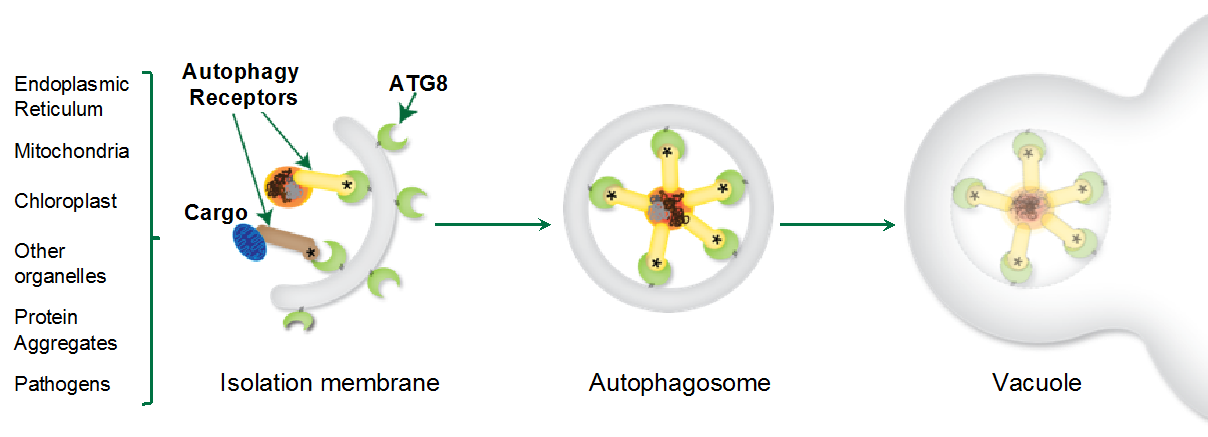
Our current research topics include:
- Exploring the contribution of ATG8 isoforms to the compartmentalization of selective autophagy responses
- Searching for novel selective autophagy receptors, ergo novel autophagy cargoes and pathways
- Searching for novel autophagy adaptors, ergo the mechanism of autophagosome trafficking and fusion with the vacuole
- Studying the evolution of autophagy pathway by comparative mechanistic approaches
Our approach:
In "What Mad Pursuit" Francis Crick says: “Classical genetics is, after all, a black-box subject. The important thing was to combine it with biochemistry. In nature hybrid species are usually sterile, but in science the reverse is often true. Hybrid subjects are often astonishingly fertile, whereas if a scientific discipline remains too pure it usually wilts.”
Following this invaluable advice, we are trying to combine genetics with biochemical, biophysical and cell biological approaches to understand the molecular principles of quality control mechanisms, specifically autophagy mediated cellular homeostasis.
To achieve our goal, through an evolutionary perspective, we are exploring three different scales of specificity:
(1)Conditional specificity. Quality control pathways will target different components under different developmental stages and stress conditions.
(2)Cell type specificity. As each cell-type has a unique response to the intrinsic and extrinsic stress factors, quality control mechanisms will also be wired in a cell-type specific manner.
(3)Subcellular specificity. Quality control pathways mostly share the same molecular players. To prevent cellular chaos, the cell must have evolved means for subcellular compartmentalization.
Model Systems:
Although not limited to, we are currently using Arabidopsis thaliana and Marchantia polymorpha as comparative model systems.
The Arabidopsis thaliana root is an excellent model system to perform mechanistic studies at cell-type specific resolution. The genetic and cell biological tools are readily established. We are complementing these tools with our autophagy toolbox to analyze spatiotemporal dynamics of autophagy at three different scales that are described above (Figure 2).
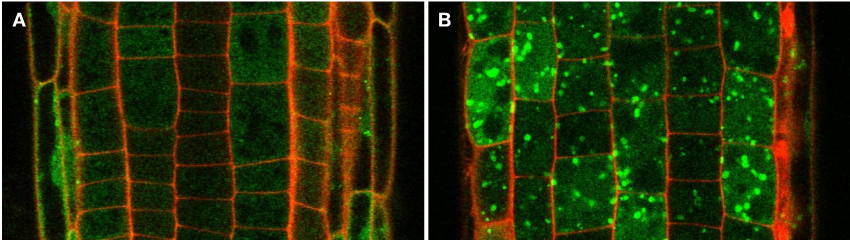
Marchantia polymorpha is an emerging model system that offers many exciting possibilities. It is easy to culture and amenable to genetic manipulation. Most importantly compared to other model systems has reduced genetic redundancy and has an extended haploid stage in its life cycle. We are particularly interested in exploiting M. polymorpha for high-throughput cell biological screens to identify novel autophagy regulators (Figure 3).

