Research
Chromatin Architecture and Function
The genetic information contained in DNA is organized into functional units assembled in specific territories within the nucleus. This organization is crucial for a proper, balanced expression of the genome. DNA is wrapped around core histone proteins H2A, H2B, H3, and H4, which form octamers the basic units of chromatin called nucleosomes. We propose that variants of the core histone proteins confer specific properties to nucleosomes and provide information that is crucial for genome organization. We focus our work on the roles of histones variants from the H3 and H2A core histones and use plants as models because they have evolved a remarkable diversity of histone variants.
H3 variants dynamics and their impacts on epigenetic marks
In recent years we have shown that H3 variants are essential in maintaining and reprogramming the epigenetic mark H3K27me3. H3K27 trimethylation is deposited by the complex Polycomb Repressive Complex 2 (PRC2) and this epigenetic mark is recognized by the complex PRC1. We uncovered that the inheritance of histone modification H3K27me3 depends on deposition of the variant H3.1, which is mediated by the chaperone CAF1 at the DNA replication fork (Jiang and Berger, 2017). These findings provide an explanation for the epigenetic memory associated with this chromatin mark.
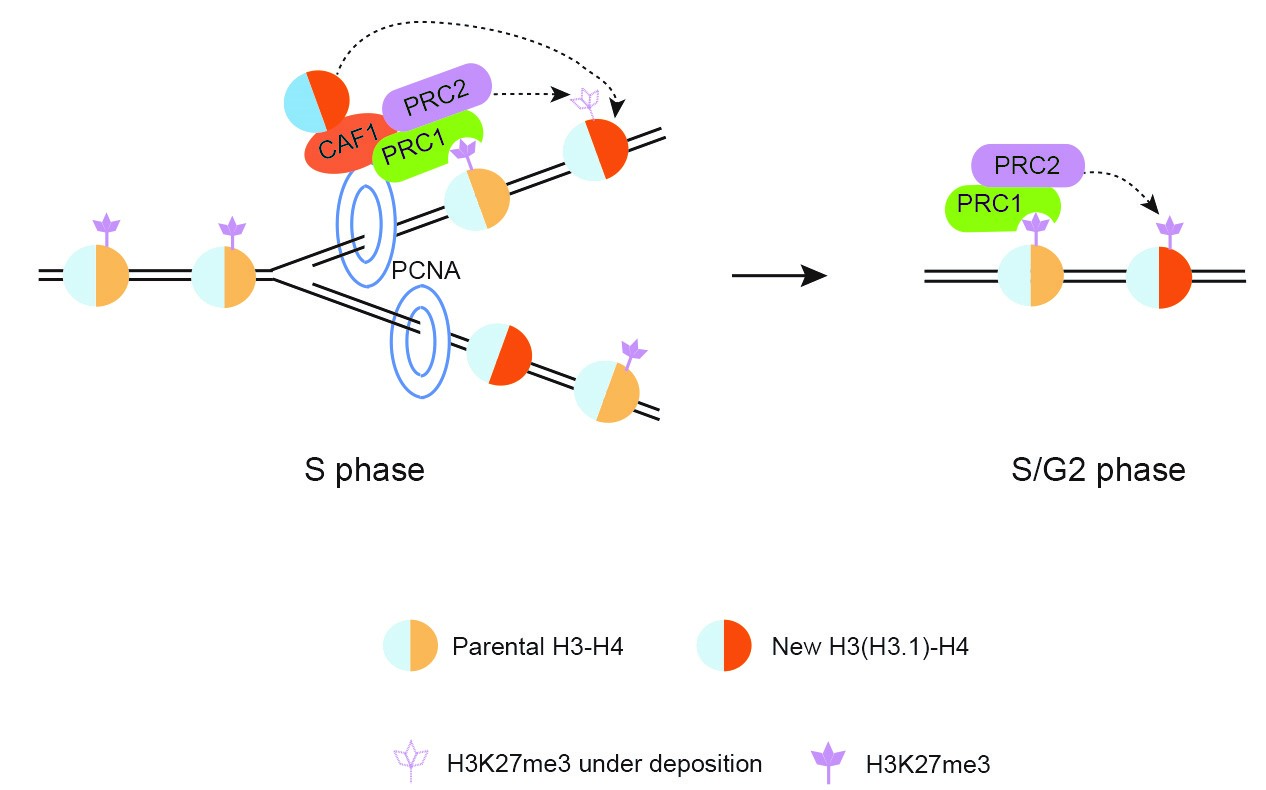
The modification H3K27me3 inhibits expression of genes that control crucial events during development. For example, the Arabidopsis gene FLC needs to be repressed during the early vegetative life and during winter to ensure flowering in spring. Because H3K27me3 is epigenetically inherited, it is essential that this repressive marks is removed at some point of the life cycle, when it is no longer required. This reprograming event has been subject to controversy. We have shown that after fertilization dynamics of exchange of non-replicative H3 variants likely participates in reprograming epigenetic marks (Ingouff et al., 2010). We are currently working on the role of a cell type specific H3 variants in reprograming H3K27me3.
H2A variants structure the chromatin landscape
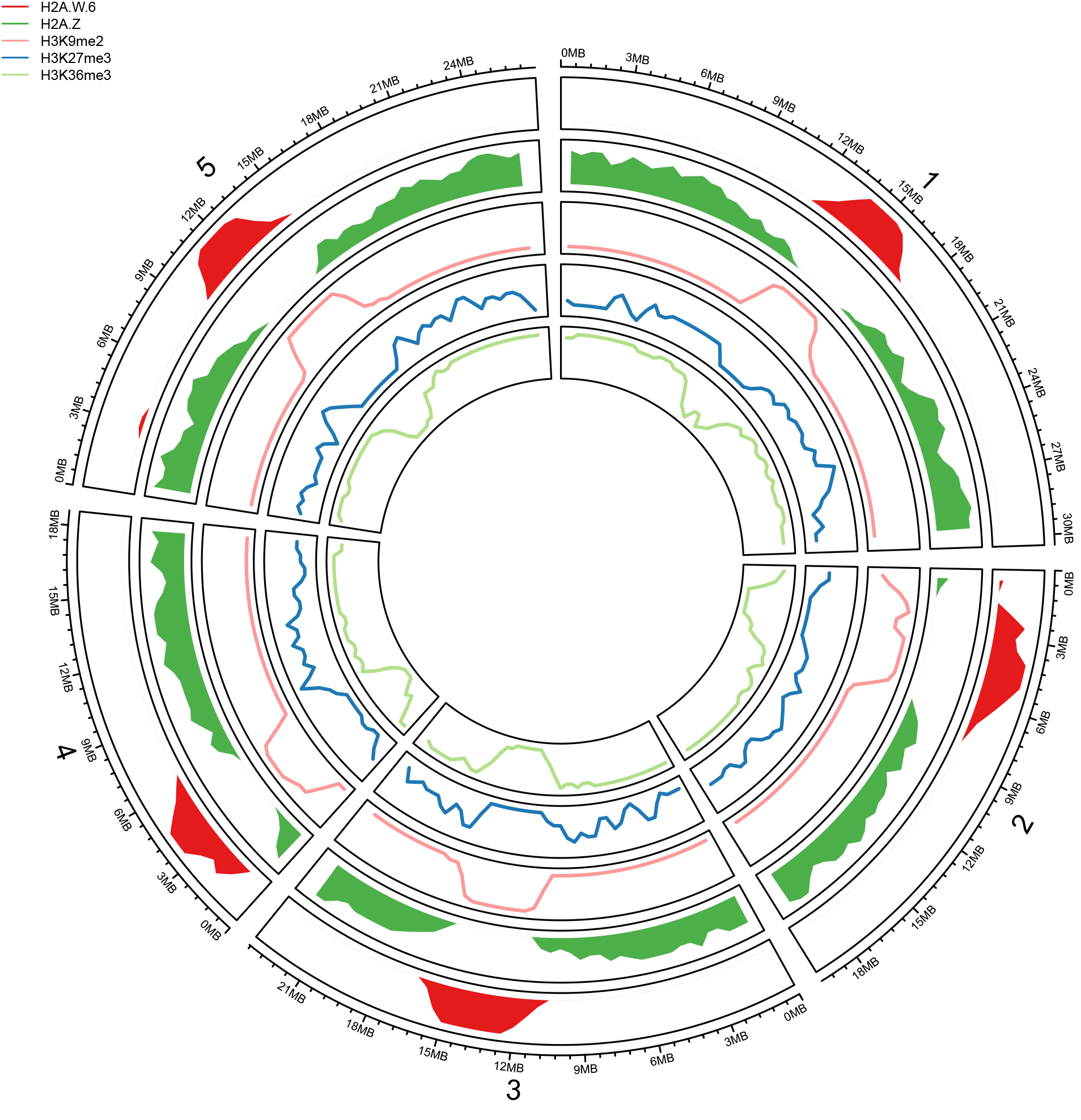
In the family of core histone H2A several variants diversified, some shared amongst all eukaryotes (H2A.Z and H2A.X) and others more specific of certain groups. Land plants evolved the specific type H2A.W, which promotes heterochromatin condensation into higher order nuclear domains (Yalegandula et al. 2014). Specific motifs in the core domain and the C-terminal tail distinguish each type of H2A variant and confer specific properties to the nucleosome (Osakabe and Lorković, 2018). We are studying the evolution, function, and mechanisms responsible for the dynamics of H2A variants using structural, biochemical, genetic and cellular approaches.
Evidence for sub-functionalization of two subtypes of H2A.W (Lorković et al., 2017) and the link between H3 variants and other epigenetic marks (Wollmann et al., 2017) led to current investigations of the roles of histone variants as major regulators of genome organization in functional units.
EvoChromo: Evolution of genome organization
We are investigating the role of essential chromatin components such as histone variants in evolution of genome organization in the plant lineage. We have obtained a chromosome-scale reassembly of the genome of Marchantia polymorpha, a species representative of early land plants (updates on http://marchantia.info/). This new resource will be used as a template to investigate how the chromatin landscape has evolved in plants. We plan to extend this work to the glaucophytes and red algae.
