
Small RNA-directed DNA elimination in Tetrahymena
Lab closed in 2016.
Transposable elements are molecular parasites that are able to move from one genome position to another. Cells in our body have a mechanism to silence these potentially harmful elements: locking transposable element into a closed form chromatin, called heterochromatin. Chemically, both transposable elements and the other parts of the genome are just stretches of DNA. So, how can cells distinguish junk from precious DNA? Evidence suggests that small RNAs, ~20-30 nucleotides in length, act as security guards to identify transposable elements. However, many remains unknown about how these small RNAs are produced, how they patrol the genome, and how they induce heterochromatin. Our group studies how short RNAs lock transposable elements into heterochromatin using the tiny-hairy protozoan Tetrahymena as a model. Moreover, Tetrahymena is able to eliminate heterochromatinized transposable elements from the genome. We are trying to understand how this special ability has been evolved.
Research
Small RNA-directed DNA elimination in Tetrahymena
Two discoveries which were awarded the Nobel Prize have shown that Tetrahymena is a useful eukaryotic model for RNA and chromatin biochemistry. Using this marvelous unicellular eukaryote as a model, we are trying to elucidate how small non-coding RNAs epigenetically regulate genome activities. For this purpose we are studying a process known as programmed DNA elimination, which is a gold mine of examples for basic biological processes, including small non-coding RNA biogenesis, selective nuclear transports, RNA and protein degradation, small and long non-coding RNA-mediated regulation of chromatin activity, chromatin organization in the nucleus, transposon silencing, and transposon-driven genome evolution.

A Tetrahymena Hsp90 co-chaperone promotes siRNA loading by ATP-dependent and ATP-independent mechanisms
The loading of small interfering RNAs (siRNAs) and microRNAs into Argonaute proteins is enhanced by Hsp90 and ATP in diverse eukaryotes. However, whether this loading also occurs independent of Hsp90 and ATP remains unclear. We found that the Tetrahymena Hsp90 co-chaperone Coi12p promotes siRNA loading into the Argonaute protein Twi1p in both, ATP-dependent and ATP-independent manners in vitro. ATP-dependent activity requires Hsp90 and the tetratricopeptide repeat (TPR) domain of Coi12p, whereas these factors are dispensable for ATP-independent activity. Both activities facilitate siRNA loading by counteracting the Twi1p-binding protein Giw1p, which is important to specifically sort the 26- to 32-nt siRNAs to Twi1p. Although Coi12p lacking its TPR domain does not bind to Hsp90, it can partially restore the siRNA loading and DNA elimination defects of COI12 knockout cells, suggesting that Hsp90- and ATP-independent loading of siRNA occurs in vivo and is an integral part of the DNA elimination process in Tetrahymena.
DNA elimination is epigenetically regulated by transnuclear genome comparison
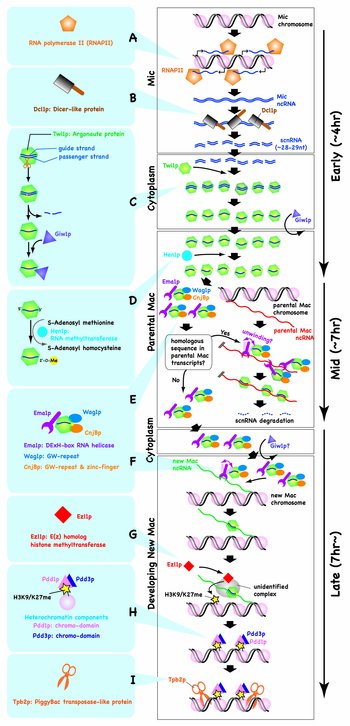
The fact that IESs do not share any common sequence motifs raises the following question: how is Tetrahymena able to identify IESs in order to induce DNA elimination? Tetrahymena solves this problem by trans-nuclear comparison of whole genomes. In a single cell, Tetrahymena has a germline known as Mic which contains complete genome including IESs, and a somatic Mac in which IESs are removed during the last sexual reproduction. Thus, the cell is able to identify IESs as sequences existing in Mic but not in Mac. Tetrahymena utilizes scnRNAs for this trans-nuclear comparison of whole genomes (Figure 2 top). This system is able to perfectly sweep away not only the existing transposons, but also any newly invaded transposons from the transcriptionally active Mac. We are trying to understand the exact molecular mechanism regulating this trans-nuclear comparison of whole genomes by small RNAs. We recently reported that only scnRNAs complementary to IESs escape degradation during conjugation, and this selective turnover of scnRNAs mediates trans-nuclear comparison of whole genomes (Figure 2, bottom). We also showed that the selective turnover of scnRNAs alone does not fully explain the observed sequence specificity of scnRNAs to IESs, because scnRNAs are produced to a greater extent from IESs than from the rest of the genome (Figure 2, lower section).
We proposed that scnRNAs target not only IESs in the new Mac for DNA elimination, but also IESs in the germline Mic to mark sites for future biased production of scnRNAs (Figure 2 top, g). Thus, DNA elimination in the new Mac may be epigenetically and transgenerationally controlled by the genome contents of the parental Mac through selective degradation of scnRNAs, and also by those of the grandparental Mac through transcriptional regulation of Mic. We believe that understanding the mechanism of DNA elimination in Tetrahymena will shed light on how ancestral genomes are able to epigenetically regulate the behavior of genomes of successive generations in general eukaryotes.
Selected Publications
(2015)
Kataoka, K., Mochizuki, K. (2015). Phosphorylation of an HP1-like Protein Regulates Heterochromatin Body Assembly for DNA Elimination. Dev Cell. 35(6):775-88
Noto, T., Kataoka, K., Suhren, JH., Hayashi, A., Woolcock, KJ., Gorovsky, MA., Mochizuki, K. (2015). Small-RNA-Mediated Genome-wide trans-Recognition Network in Tetrahymena DNA Elimination. Mol Cell. 59(2):229-42
Woehrer, SL., Aronica, L., Suhren, JH., Busch, CJ., Noto, T., Mochizuki, K. (2015). A Tetrahymena Hsp90 co-chaperone promotes siRNA loading by ATP-dependent and ATP-independent mechanisms. EMBO J. 34(4):559-77
(2013)
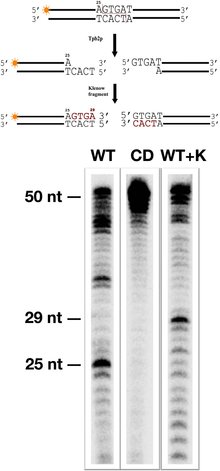
Vogt, A., Mochizuki, K. (2013). A domesticated PiggyBac transposase interacts with heterochromatin and catalyzes reproducible DNA elimination in Tetrahymena. PLoS Genet. 9(12):e1004032
(2012)
Schoeberl, UE., Kurth, HM., Noto, T., Mochizuki, K. (2012). Biased transcription and selective degradation of small RNAs shape the pattern of DNA elimination in Tetrahymena. Genes Dev. 26(15):1729-42
(2010)
Noto, T., Kurth, HM., Kataoka, K., Aronica, L., DeSouza, LV., Siu, KW., Pearlman, RE., Gorovsky, MA., Mochizuki, K. (2010). The Tetrahymena argonaute-binding protein Giw1p directs a mature argonaute-siRNA complex to the nucleus. Cell. 140(5):692-703
(2009)
Kurth, HM., Mochizuki, K. (2009). 2'-O-methylation stabilizes Piwi-associated small RNAs and ensures DNA elimination in Tetrahymena. RNA. 15(4):675-85
(2008)
Aronica, L., Bednenko, J., Noto, T., DeSouza, LV., Siu, KW., Loidl, J., Pearlman, RE., Gorovsky, MA., Mochizuki, K. (2008). Study of an RNA helicase implicates small RNA-noncoding RNA interactions in programmed DNA elimination in Tetrahymena. Genes Dev. 22(16):2228-41