Research
Population Genetics
The 1001 Genomes Project
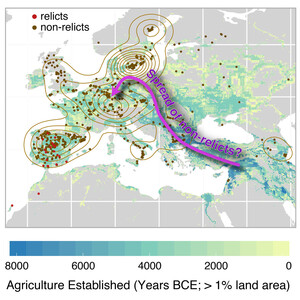
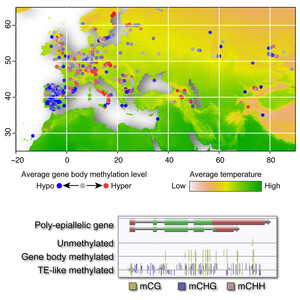
Thanks to decreasing genotyping costs, there is currently great interest in so-called genome-wide association studies (GWAS), in which one attempts to identify genes responsible for variation simply by correlating genotype (typically in the form of single nucleotide polymorphisms) with phenotype. The model plant A. thaliana is ideally suited for such studies because it naturally occurs as inbred lines which can be genotyped once and phenotyped repeatedly. For over 15 years, we have been spearheading an international effort to make genome-wide association in A. thaliana a reality, and we recently published the genomes, transcriptomes, and epigenomes of over 1000 natural inbred lines — a fantastic resource for the genetic community. We are also supporting public websites and databases that allow anyone to carry out GWAS and help coordinate as much phenotypic information. More information on the 1001 Genotypes project can be found on the project website, 1001genomes.org/.
The genetics of adaptation


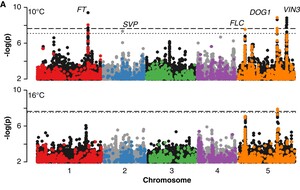
We are carrying out large-scale GWAS to understand the genetic basis of variation for adaptively important traits like flowering time, dormancy, and cold tolerance. The GWAS results are complemented with a variety of methods to confirm results. Our goal is to achieve as complete an understanding of the genetics of these traits as is possible. Investigating the adaptive significance of any trait also requires field studies. We are using field sites in northern and southern Sweden for reciprocal transplant competition experiments of both natural inbred lines and the offspring of crosses. The objective is to map the genes responsible for fitness differences, and to characterize them at the molecular level.
Speciation in the genus Arabidopsis
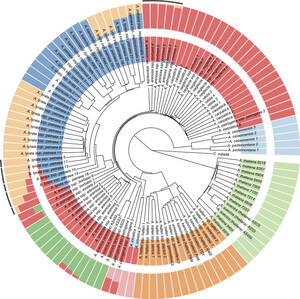
Differences between individuals within species are micro-versions of differences between species. Understanding the nature of species differences, and the process by which they arise, is a long-standing question in biology — one that modern DNA sequencing methods allow us to tackle using brute force. We are doing this is in several groups of organisms, one of them being the genus Arabidopsis, home of the model plant A. thaliana. Long-term questions include the evolution of genome size, the effects of polyploidy, and the switch to self-fertilization, but our immediate goal was to understand how genetic variation is distributed across a diverse group of plant species. To this end, we sequenced over one-hundred individuals from all taxa in the genus, and demonstrated that speciation in the genus is a messy (and ongoing) process involving long periods of partial reproductive isolation.
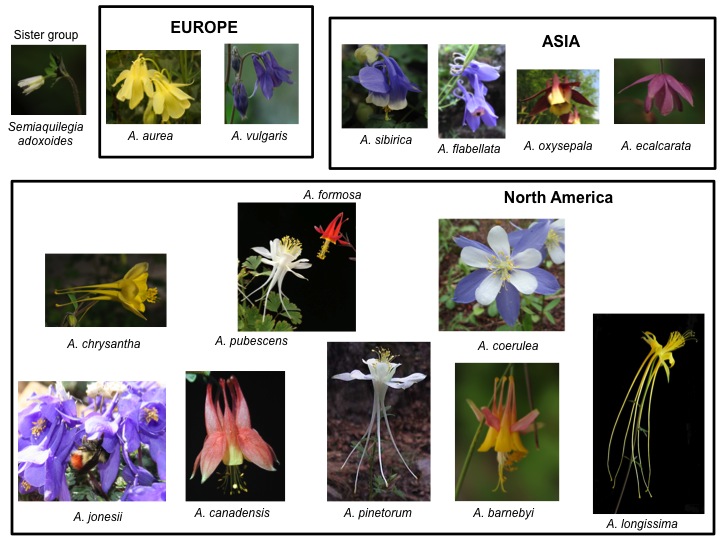
Speciation in Aquilegia
We are also studying the genetics of species differences in the columbine genus, Aquilegia (Ranunculaceae). The genus is a beautiful example of a recent, rapid, adaptive radiation, especially with respect to floral morphology and color. We seek to understand the genetic basis for such striking differences, focusing particularly on two North American species, A. formosa and A. pubescens, the former of which is pollinated by hummingbirds, the latter of which is pollinated by hawkmoths, resulting in reproductive isolation. However, the two species are completely inter-fertile and form natural hybrid zones. We have demonstrated that the two species are very closely related at the genetic level, with most polymorphisms shared between the species, and little divergence in allele frequencies, and we are now trying to identify the genes responsible for the phenotypic differences through GWAS.
Speciation in African green monkeys

The African green monkey (Cercopithecus sp.) is a common Old World monkey, spread throughout much of Africa, and introduced by humans to the Caribbean. It is also kept in large colonies for behavioral and biomedical research, in particular for understanding HIV resistance. As part of an international consortium to develop genomic resources for vervets, we have sequenced over 100 monkeys sampled across the African continent, covering all known species, and discovered dramatic footprints of selection at genes involved in virus response.
